|
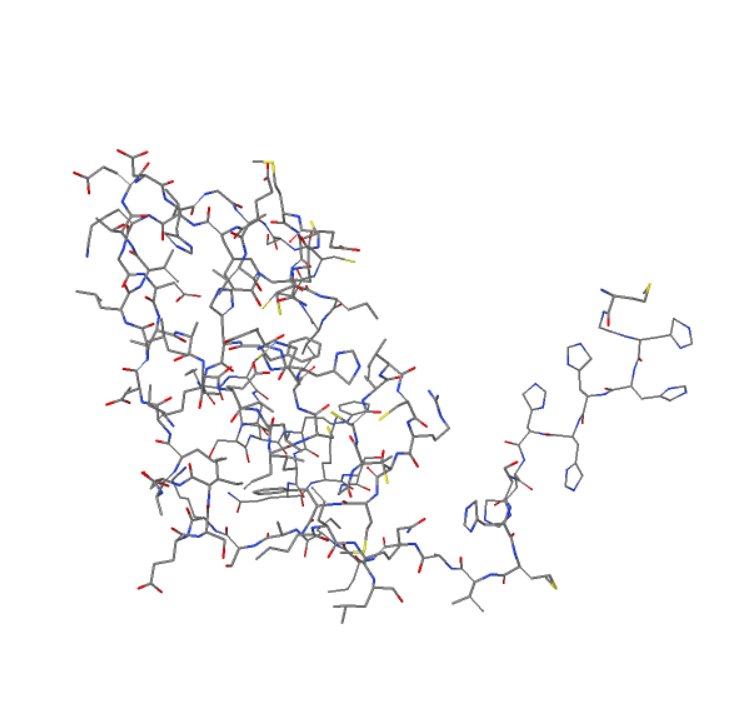
BetaSCPWeb predicts the optimal side-chain conformation of protein structure
using the input of an amino acid sequence and a backbone conformation
and reports the predicted structure output as a visual, a textual, and a PDB file formats.
Learn more... |
|
* Citing BetaSCPWeb*
J. Ryu, M. Lee, J. Cha, R. Laskowski, S.-E. Ryu, D.-S. Kim,
BetaSCPWeb: side-chain prediction for protein structures using Voronoi diagrams and geometry prioritization,
Nucleic Acids Research, (doi: 10.1093/nar/gkw368), 2016.